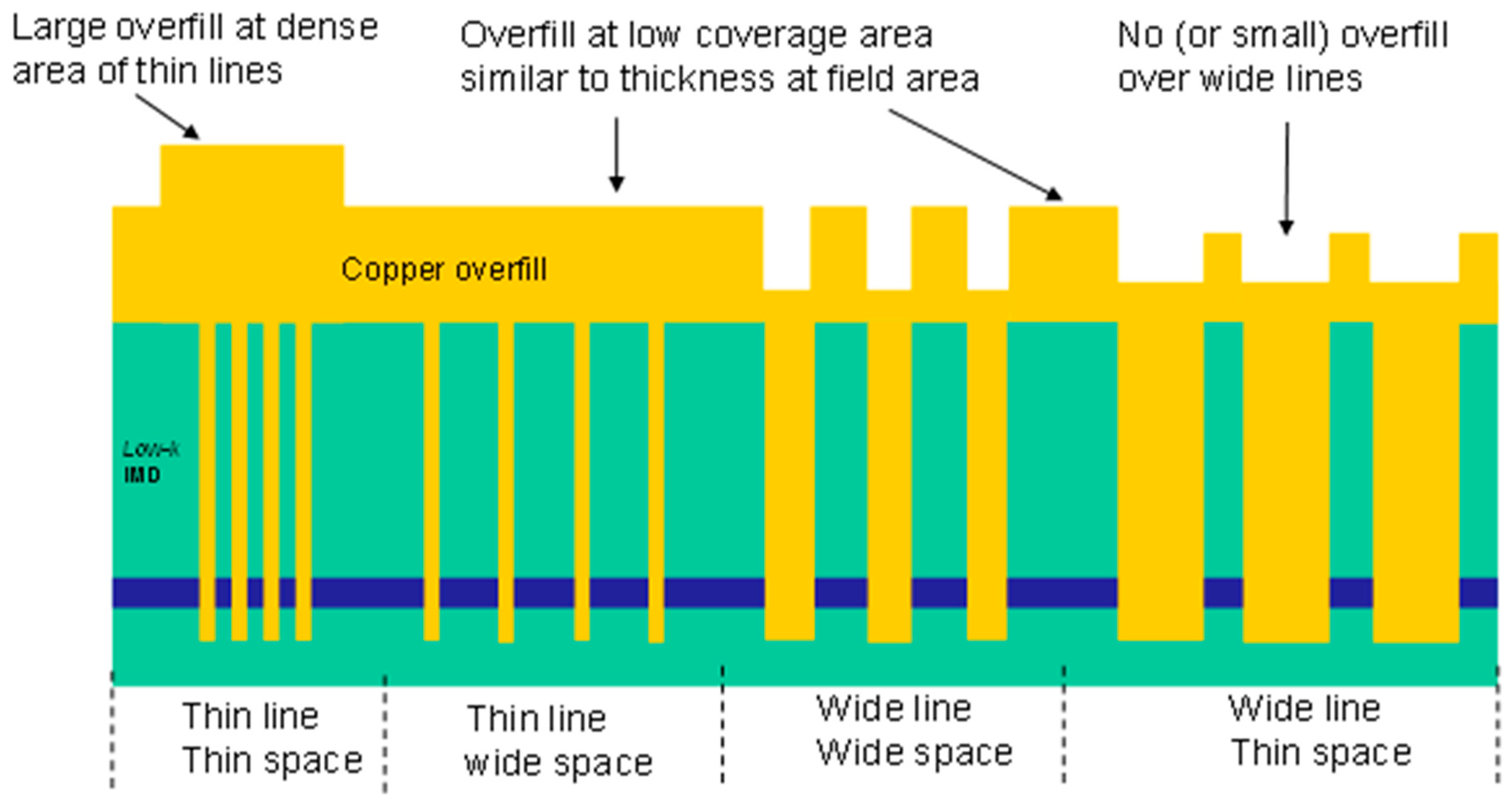
The variables for NGS experiments: coverage, read length, multiplexing
4.6 (479) · $ 12.50 · In stock
Basics of NGS, Target Capture
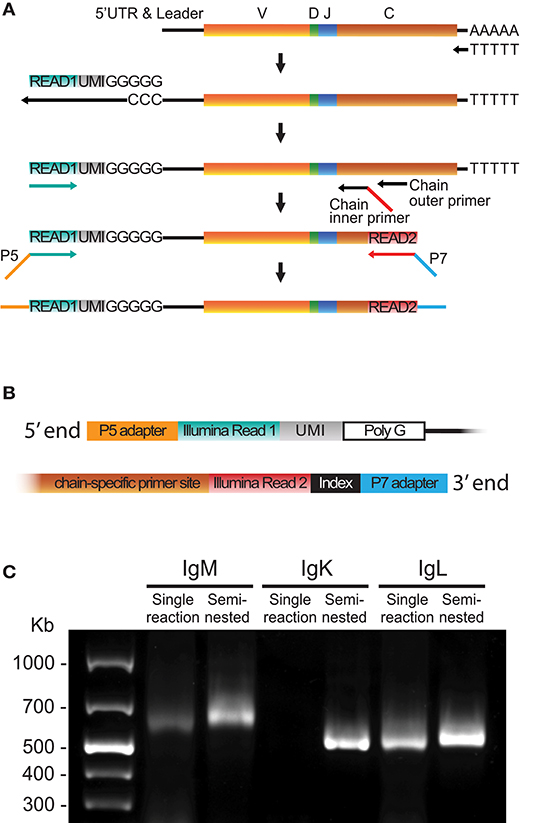
Frontiers High-Quality Library Preparation for NGS-Based Immunoglobulin Germline Gene Inference and Repertoire Expression Analysis
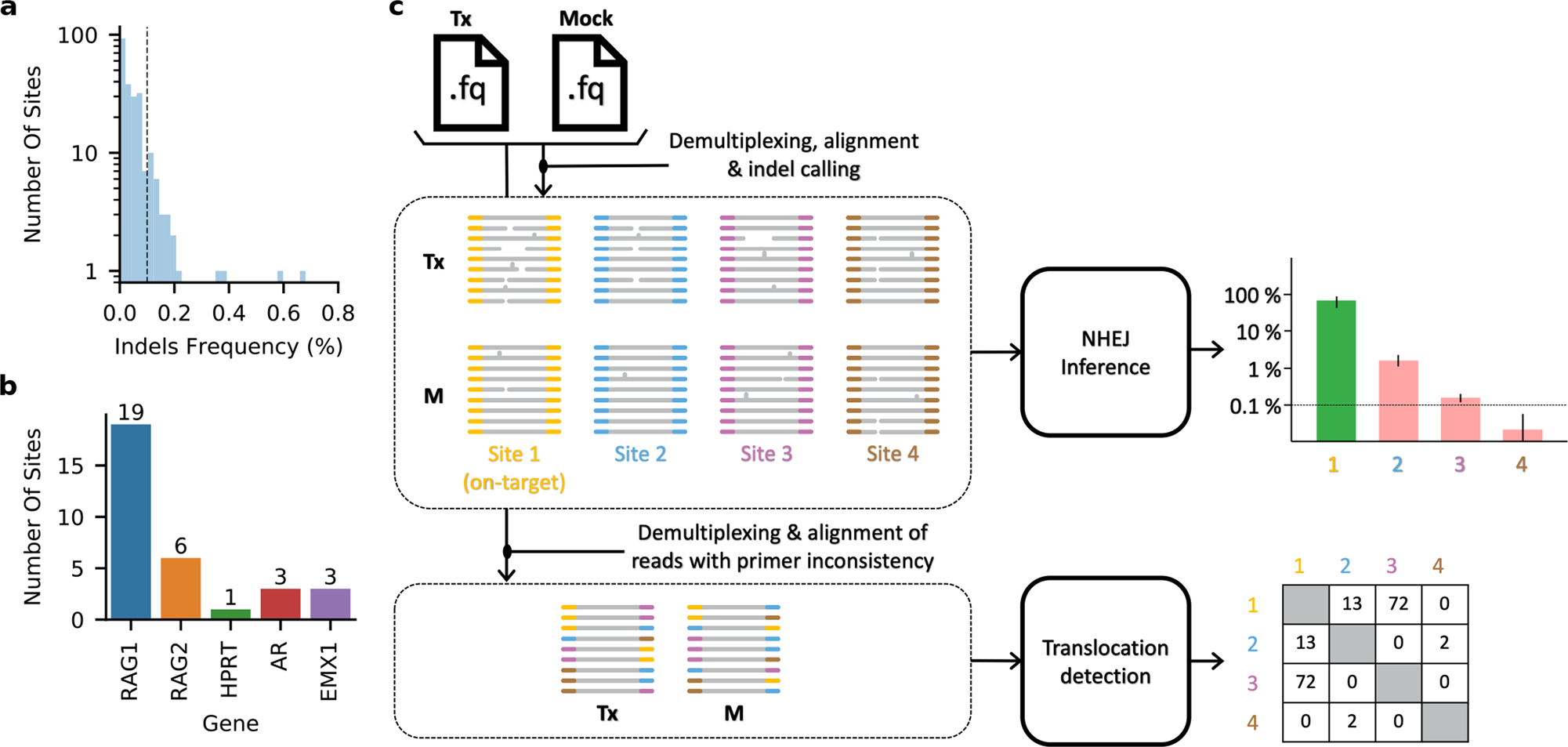
CRISPECTOR provides accurate estimation of genome editing translocation and off-target activity from comparative NGS data
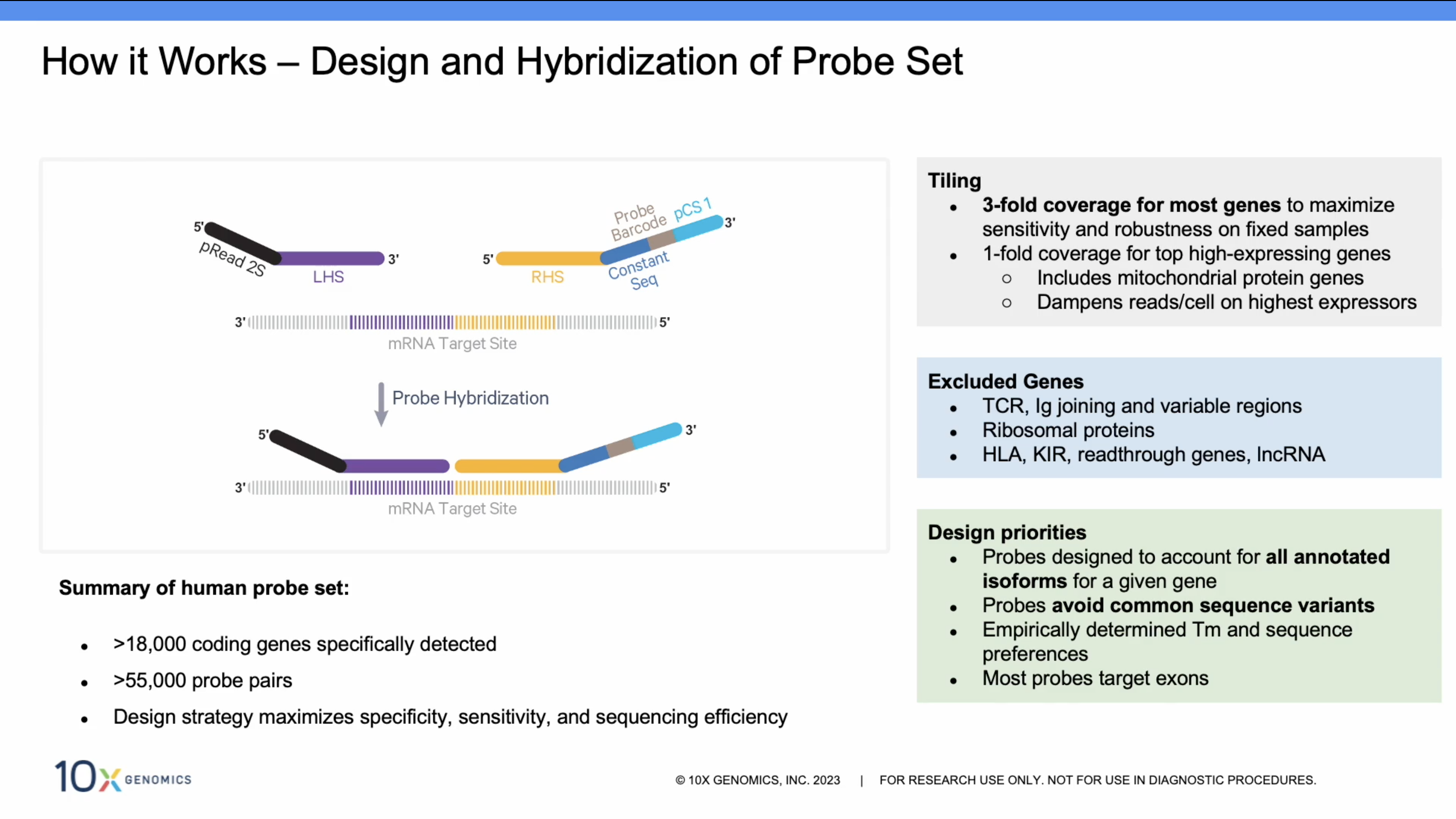
Single Cell Gene Expression Flex - 10x Genomics

NGS - Experimental Design
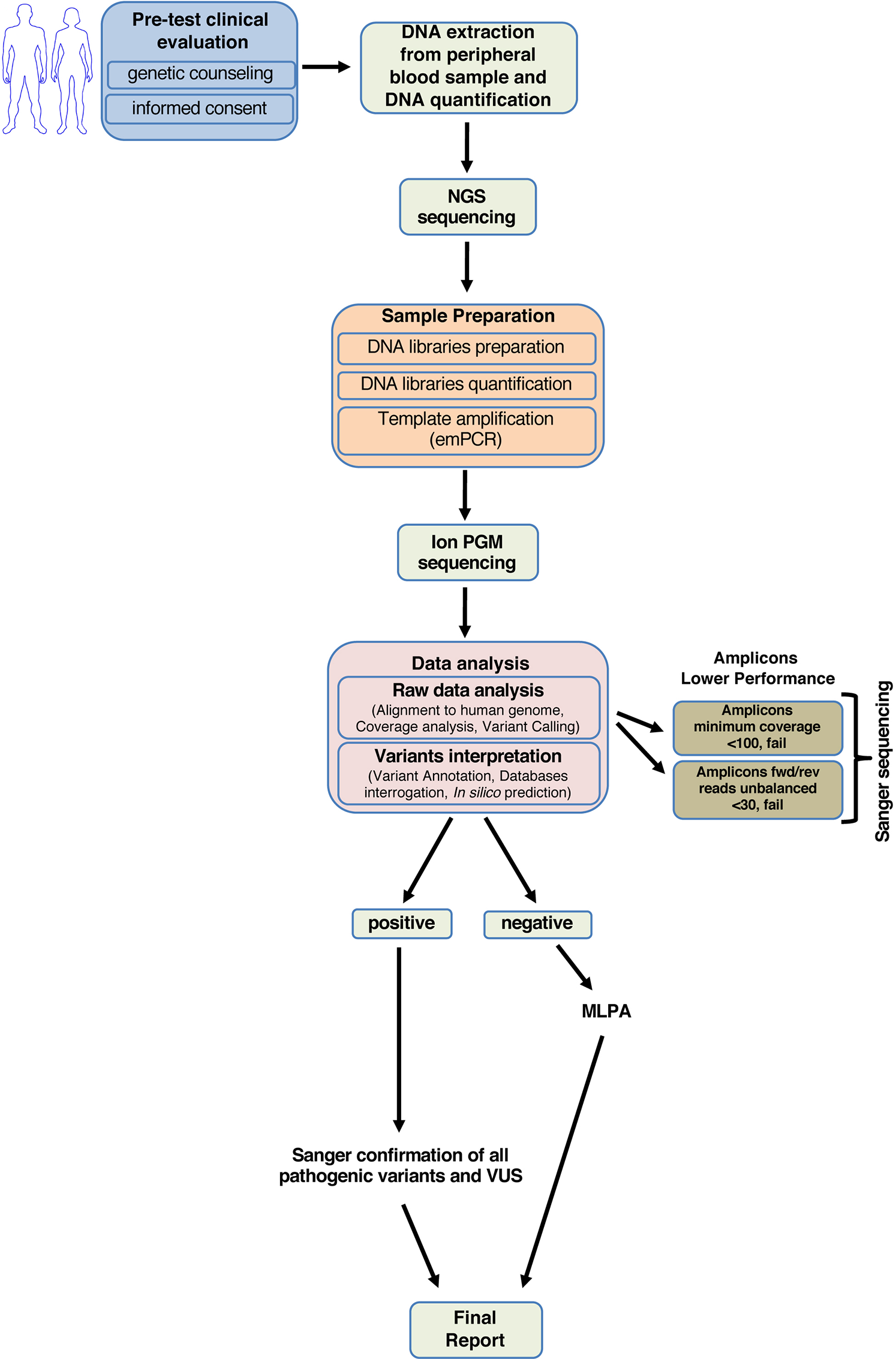
Next-generation sequencing of BRCA1 and BRCA2 genes for rapid detection of germline mutations in hereditary breast/ovarian cancer [PeerJ]

Highlighting Clinical Metagenomics for Enhanced Diagnostic Decision-making: A Step Towards Wider Implementation - Computational and Structural Biotechnology Journal
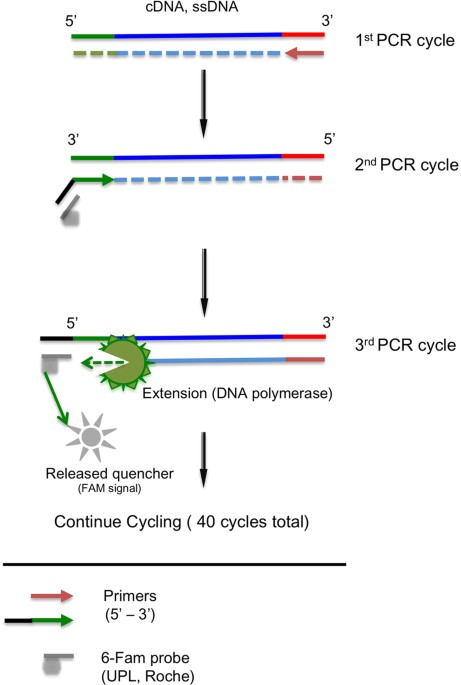
Comparison of DNA Quantification Methods for Next Generation Sequencing

What is mappable coverage?
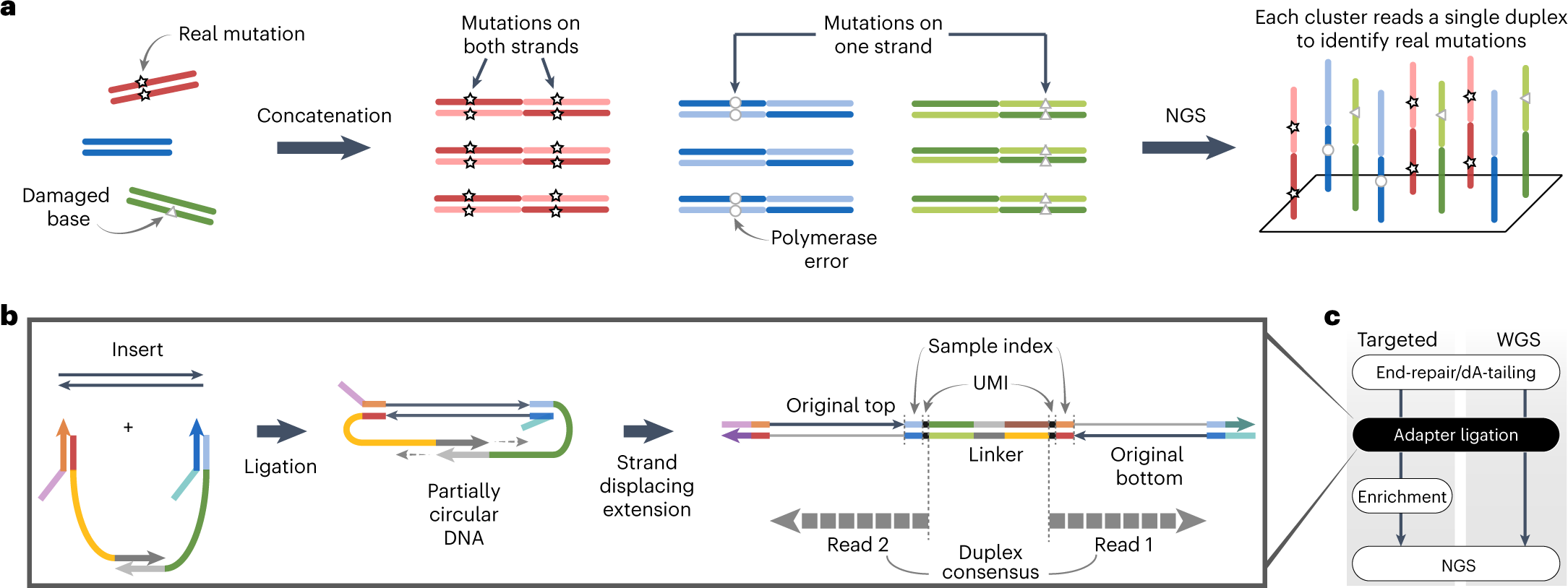
Single duplex DNA sequencing with CODEC detects mutations with high sensitivity

Identifying the best PCR enzyme for library amplification in NGS

NASCarD (Nanopore Adaptive Sampling with Carrier DNA): A rapid, PCR-free method for whole genome sequencing of pathogens in clinical samples
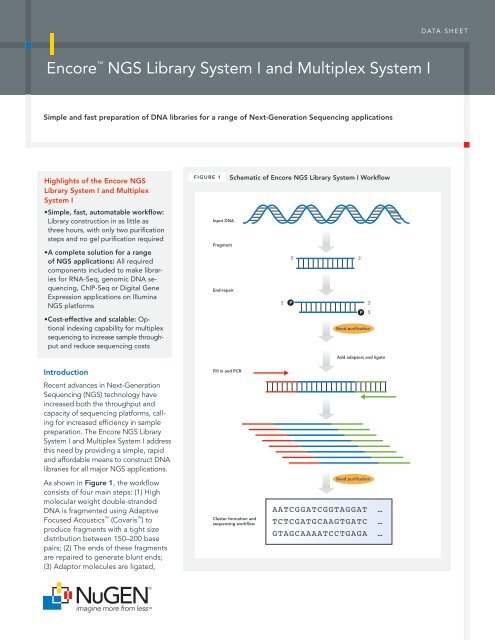
encore™ NGS Library System I and Multiplex System I
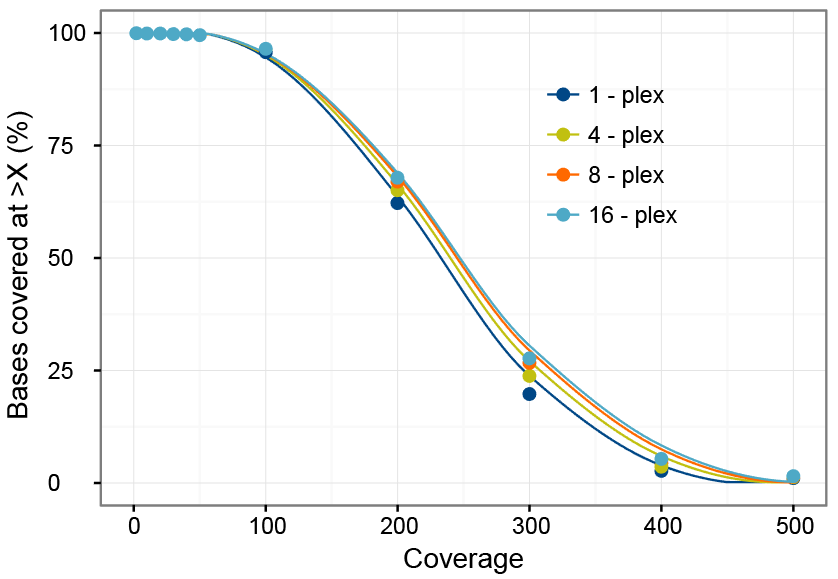
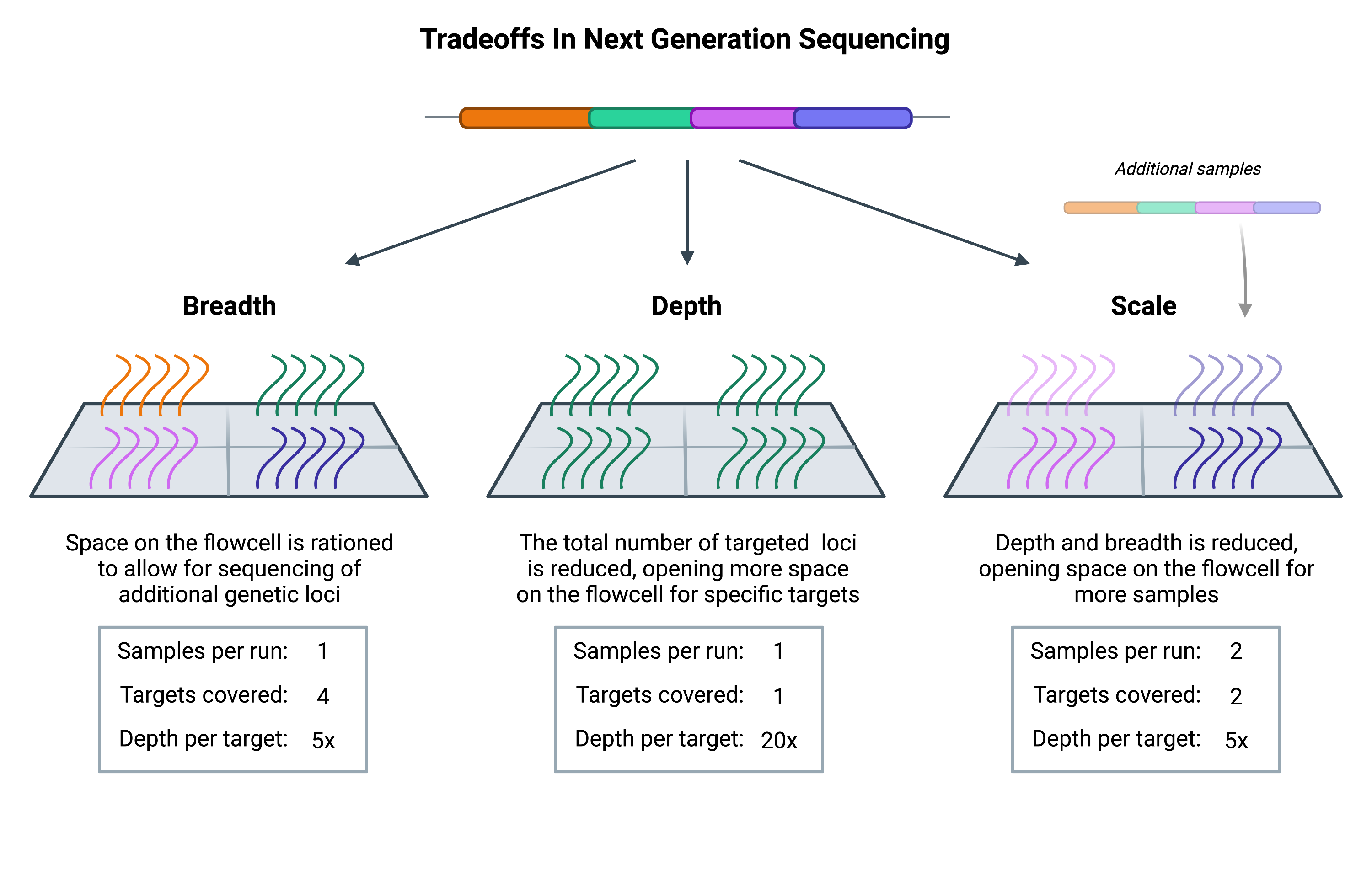
Multiplexed targeted next generation sequencing coverage

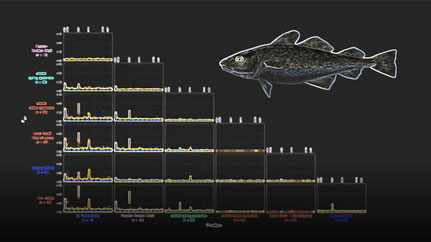
Depth of Coverage: The histograms depict the read depth by all called